LATEST RESEARCH
Chemical engineering of CRISPR-Cas systems for therapeutic application
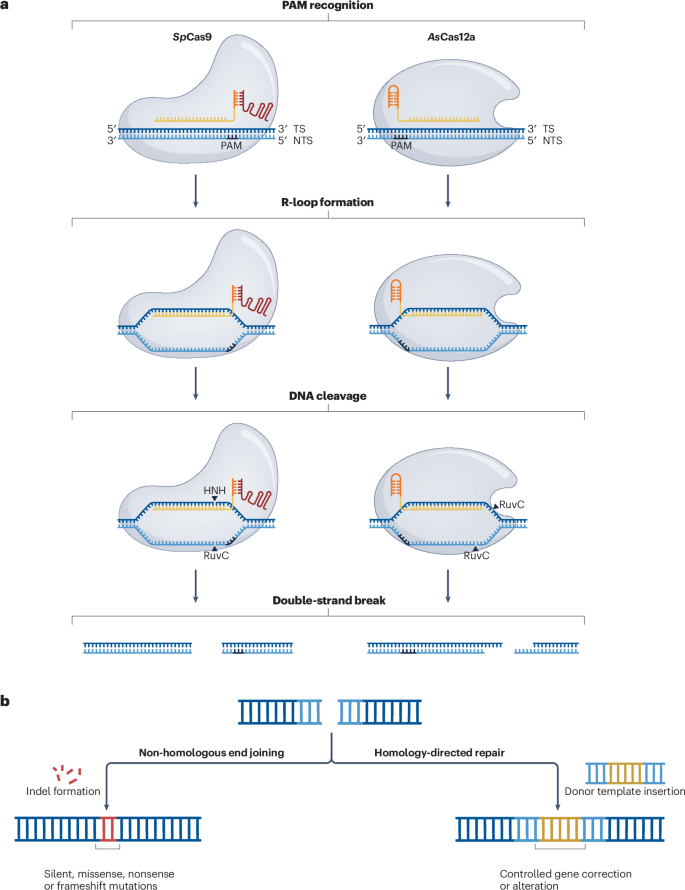
Our collaboration with the Gagnon group at Wake Forest University continues! Following on from the amazing work investigating the modification of the pseudoknot of Cas12a's crRNA (available here), this detailed review covers the advancements made in the chemical engineering of CRISPR-Cas systems over the last few years, and the future of the newest generation of RNA therapeutics.
H.M. Barber, A.A. Pater, K.T. Gagnon†, M. J. Damha†, and D. O'Reilly† (2024) Chemical engineering of CRISPR-Cas systems for therapeutic application, Nat. Rev. Drug Discov., doi:10.1038/s41573-024-01086-0
View the full article here.
"Milling and Shaking" process to make DNA fragments
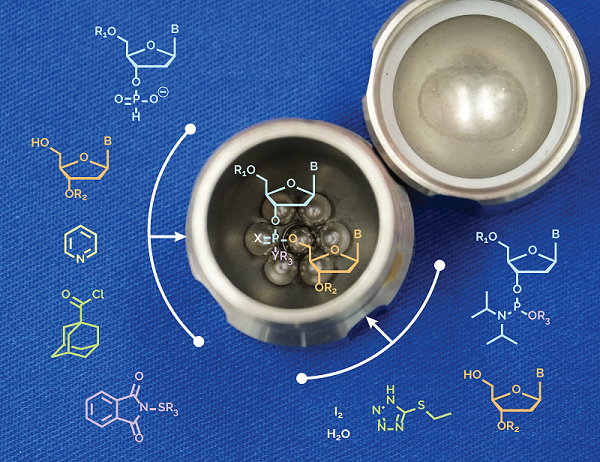
Our students James Thorpe and Daniel O’Reilly joined forces to demonstrate that di‐ and trinucleotides can be assembled under ball‐milling conditions by multi‐step reactions, in a one‐pot fashion, and in the absence of bulk solvents. The process is fully compatible with H‐phosphonate and phosphoramidite chemistries, currently applied to oligo‐DNA and RNA synthesis.
J.D. Thorpe, D. O’Reilly, T. Friščić†, and M. J. Damha† (2020) Mechanochemical synthesis of short DNA fragments, Chem. Eur. J. 26, 8857-8861.
View the full article here.
Substrate analogs that trap the 2′-phospho-ADP-ribosylated RNA intermediate of the Tpt1 (tRNA 2′-phosphotransferase) reaction pathway
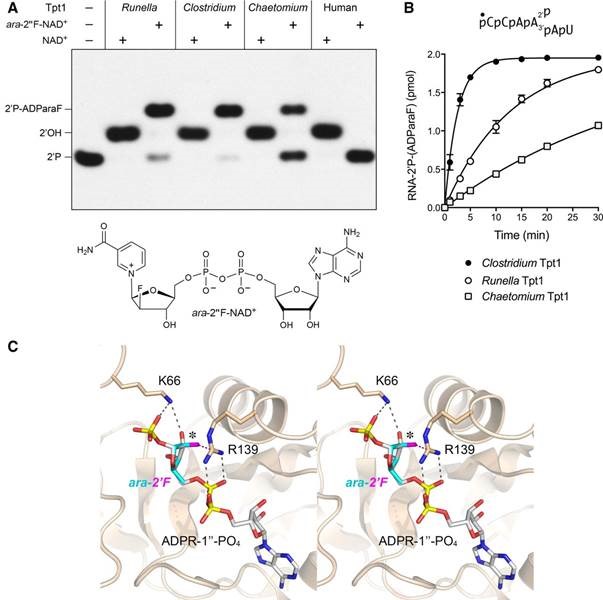
Leonora Abdullahu from our group teamed up with Dr. Stewart Shuman and co-workers (Sloan-Kettering Institute, New York) to study Tpt1 - an essential enzyme in the fungal and plant tRNA splicing pathways that removes the 2′-PO4 at the splice junction generated by fungal and plant tRNA ligases. Leonora is currently developing inhibitors of the enzyme as a potential approach to develop novel antifungal agents and to obtain crystallographic structures of Tpt1.
S. Dantuluri, L. Abdullahu, A. Munir, A. Katolik, M.J. Damha†, and S. Shuman† (2020) Substrate analogs that trap the 2'-phospho-ADP-ribosylated RNA intermediate of the Tpt1 (tRNA 2'-phosphotransferase) reaction pathway, RNA 26, 373-381. doi: 10.1261/rna.074377.119
View the full article here.
2′-Fluoroarabinonucleic acid modification traps G-quadruplex and i-motif structures in human telomeric DNA
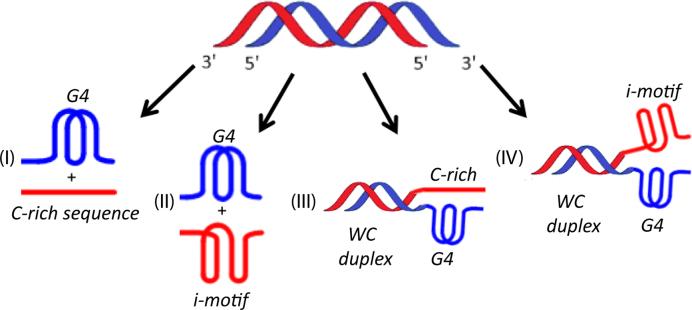
Human telomeres and promoter regions of genes fulfill a significant role in cellular aging and cancer. These regions comprise of guanine and cytosine-rich repeats, which under certain conditions can fold into G-quadruplex (G4) and i-motif structures, respectively. Herein, we use UV, circular dichroism and NMR spectroscopy to study several human telomeric sequences and demonstrate that G4/i-motif-duplex interconversion kinetics are slowed down dramatically by 2′-β-fluorination and the presence of G4/i-motif-duplex junctions. NMR-monitored kinetic experiments on 1:1 mixtures of native and modified C- and G-rich human telomeric sequences reveal that strand hybridization kinetics are controlled by G4 or i-motif unfolding. Furthermore, we provide NMR evidence for the formation of a hybrid complex containing G4 and i-motif structures proximal to a duplex DNA segment at neutral pH. While the presence of i-motif and G4 folds may be mutually exclusive in promoter genome sequences, our results suggest that they may co-exist transiently as intermediates in telomeric sequences.
H. Abou Assi, C. Gonzalez,† and M.J. Damha† (2017). 2'-Fluoroarabinonucleic acid modification traps G-quadruplex and i-motif structures in human telomeric DNA. Nucleic Acids Research, doi: 10.1093/nar/gkx838
View the full article here.
Mapping the Affinity Landscape of Thrombin-Binding Aptamers on 2'F-ANA/DNA Chimeric G-Quadruplex Microarrays
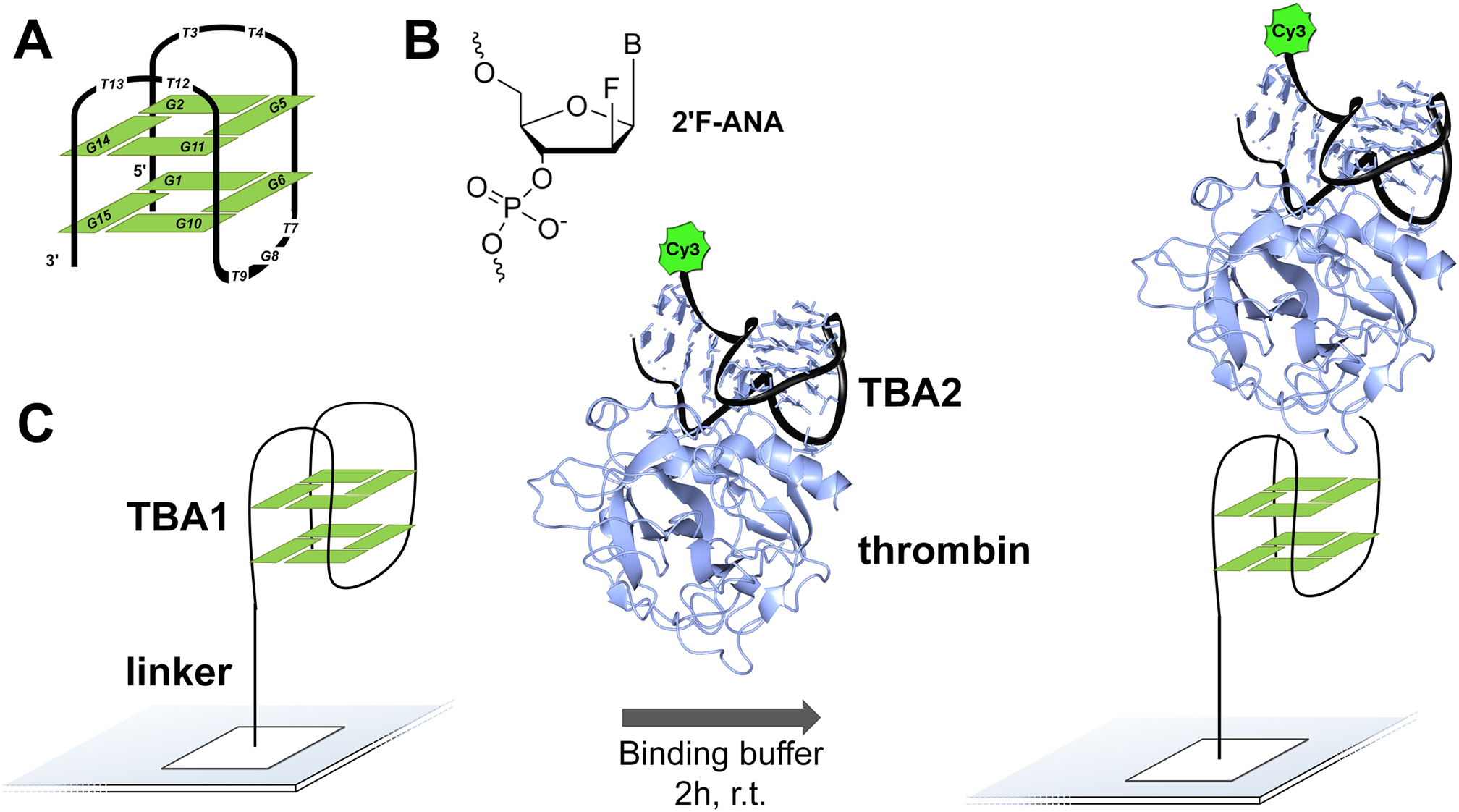
In situ fabricated nucleic acids microarrays are versatile and very high-throughput platforms for aptamer optimization and discovery, but the chemical space that can be probed against a given target has largely been confined to DNA, while RNA and non-natural nucleic acid microarrays are still an essentially uncharted territory. 2΄-Fluoroarabinonucleic acid (2΄F-ANA) is a prime candidate for such use in microarrays. Indeed, 2΄F-ANA chemistry is readily amenable to photolithographic microarray synthesis and its potential in high affinity aptamers has been recently discovered. We thus synthesized the first microarrays containing 2΄F-ANA and 2΄F-ANA/DNA chimeric sequences to fully map the binding affinity landscape of the TBA1 thrombin-binding G-quadruplex aptamer containing all 32 768 possible DNA-to-2΄F-ANA mutations. The resulting microarray was screened against thrombin to identify a series of promising 2΄F-ANA-modified aptamer candidates with Kds significantly lower than that of the unmodified control and which were found to adopt highly stable, antiparallel-folded G-quadruplex structures. The solution structure of the TBA1 aptamer modified with 2΄F-ANA at position T3 shows that fluorine substitution preorganizes the dinucleotide loop into the proper conformation for interaction with thrombin. Overall, our work strengthens the potential of 2΄F-ANA in aptamer research and further expands non-genomic applications of nucleic acids microarrays.
J. Lietard†, H. Abou-Assi, I. Gomez-Pinto, C. Gonzalez, M.M. Somoza and M.J. Damha† (2017) Mapping the affinity landscape of thrombin-binding aptamers on 2'F-ANA/DNA chimeric G-quadruplex microarrays. Nucleic Acids Research, 45: 1619–1632, doi: 10.1093/nar/gkw1357
View the full article here.
Fluorescent Branched RNAs for High-Throughput Analysis of Dbr1 Enzyme Kinetics and Inhibition
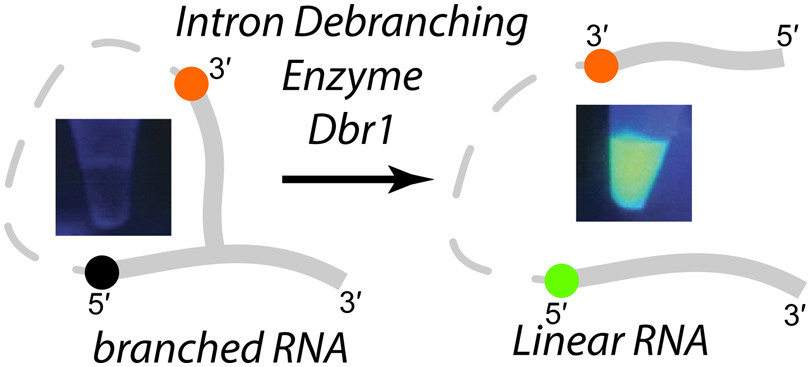
We have developed fluorescent 2′,5′ branched RNAs (bRNA) that permit real time monitoring of RNA lariat (intron) debranching enzyme (Dbr1) kinetics. These compounds contain fluorescein (FAM) on the 5′ arm of the bRNA that is quenched by a dabcyl moiety on the 2′ arm. Dbr1-mediated hydrolysis of the 2′,5′ linkage induces a large increase in fluorescence, providing a convenient assay for Dbr1 hydrolysis. We show that unlabeled bRNAs with non-native 2′,5′-phosphodiester linkages, such as phosphoramidate or phosphorothioate, can inhibit Dbr1-mediated debranching with IC50 values in the low nanomolar range. In addition to measuring kinetic parameters of the debranching enzyme, these probes can be used for high throughput screening (HTS) of chemical libraries with the aim of identifying Dbr1 inhibitors, compounds that may be useful in treating neurodegenerative diseases and retroviral infections.
A. Katolik, N.E. Clark†, N. Tago, E.J. Montemayor, P.J. Hart†, and M.J. Damha† (2017) Fluorescent branched RNAs for high-throughput analysis of Dbr1 enzyme kinetics and inhibition. ACS Chemical Biology, 12: 622–627, doi: 10.1021/acschembio.6b00971
View the full article here.
Stabilization of i-Motif Structures by 2'-B-Fluorination of DNA
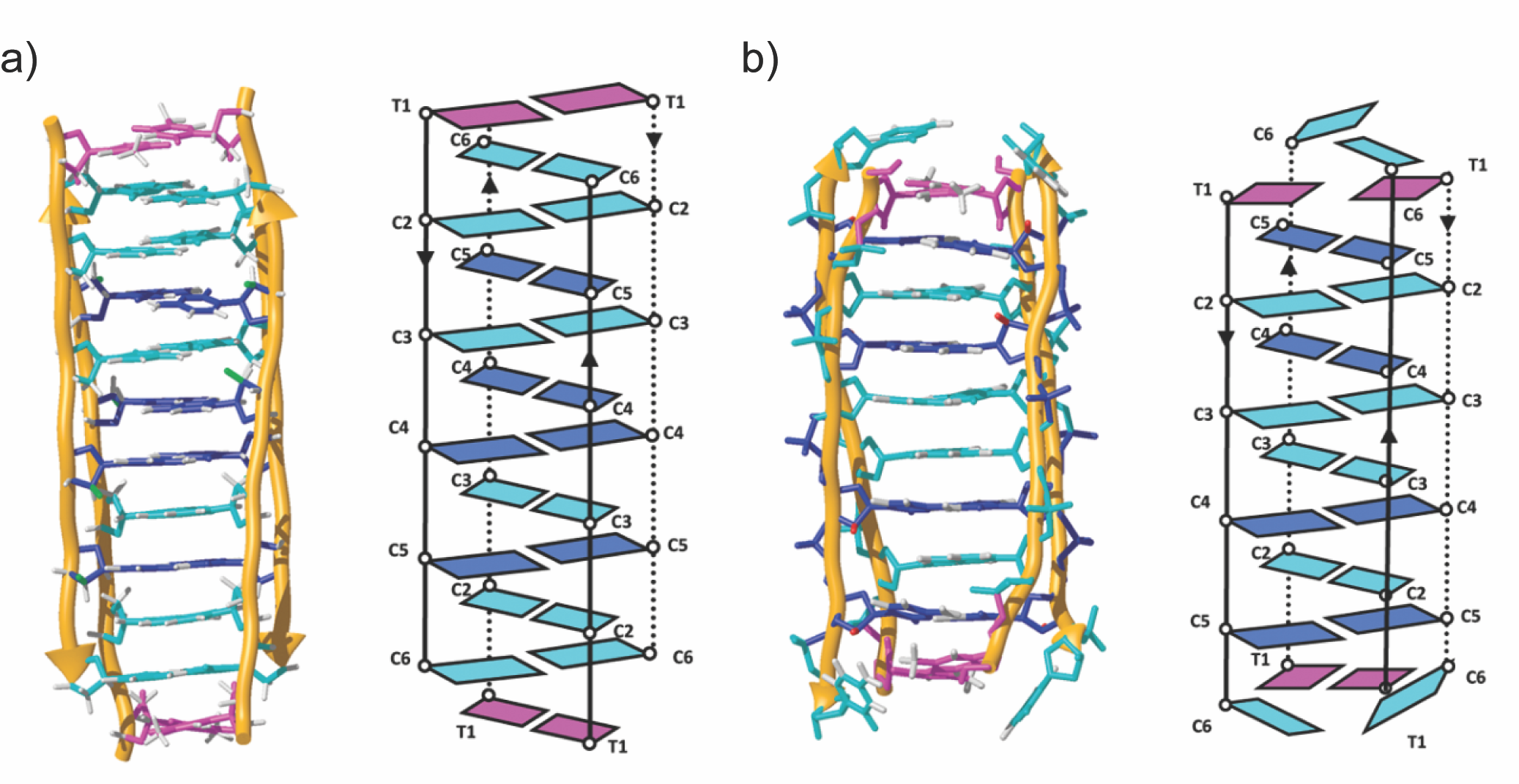
This publication was recommended on F1000Prime by Jean-Louis Mergny, a Research Director in Structural Biology at the Université de Bordeaux. Congratulations to Hala!
i-Motifs are four-stranded DNA structures consisting of two parallel DNA duplexes held together by hemi-protonated and intercalated cytosine base pairs (C:CH+). They have attracted considerable research interest for their potential role in gene regulation and their use as pH responsive switches and building blocks in macromolecular assemblies. At neutral and basic pH values, the cytosine bases deprotonate and the structure unfolds into single strands. To avoid this limitation and expand the range of environmental conditions supporting i-motif folding, we replaced the sugar in DNA by 2-deoxy-2-fluoroarabinose. We demonstrate that such a modification significantly stabilizes i-motif formation over a wide pH range, including pH 7. Nuclear magnetic resonance experiments reveal that 2-deoxy-2-fluoroarabinose adopts a C2′-endo conformation, instead of the C3′-endo conformation usually found in unmodified i-motifs. Nevertheless, this substitution does not alter the overall i-motif structure. This conformational change, together with the changes in charge distribution in the sugar caused by the electronegative fluorine atoms, leads to a number of favorable sequential and inter-strand electrostatic interactions. The availability of folded i-motifs at neutral pH will aid investigations into the biological function of i-motifs in vitro, and will expand i-motif applications in nanotechnology.
H. Abou Assi, R.W. Harkness V, N. Martín-Pintado, C.J. Wilds, R. Campos-Olivas, A.K. Mittermaier, C. González†, and M.J. Damha† (2016) Stabilization of i-motif structures by 2'-β-fluorination of DNA , Nucleic Acids Research. doi: 10.1093/nar/gkw402.
View the full article here.
Synthesis and Studies of Oxepane Nucleoside Analogues
The synthesis of a novel series of seven-membered ring nucleoside analogues as candidates for biological screening and gene silencing applications is described. The key step in the synthetic approach is a stereoselective synthesis of an epoxide that is used as a common synthetic intermediate to prepare functionalized oxepane nucleoside derivatives. The conformational landscape and preferred ring-puckering of selected oxepane nucleosides was also studied by NMR, X-ray crystallography, and quantum mechanical calculations.
M. Habibian, S. Martinez-Montero, G. Portella, Z. Chua, D.S. Bohle, M. Orozco, and M.J. Damha† (2015) Seven-membered ring nucleoside analogues: stereoselective synthesis and studies on their conformational properties, Organic Letters, 17 (21): 5416-5419.
View the full article here.
Nucleotide Sugar Pucker Preference Mitigates Excision by HIV-1 RT
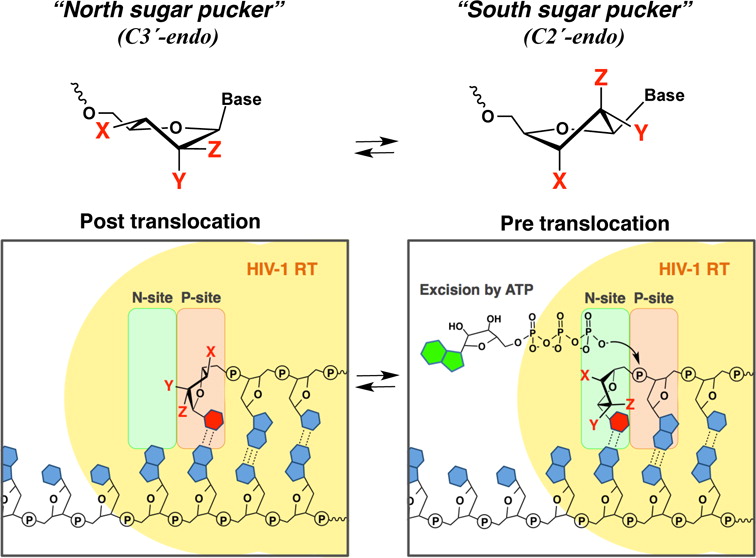
A series of DNA primers containing nucleotides with various sugar pucker conformations at the 3′-terminus were chemically synthesized by solid-phase synthesis. The ability of wild-type (WT) HIV-1 reverse transcriptase (RT) and AZT-resistant (AZTr) RT to excise the 3′-terminal nucleotide was assessed. Nucleosides with a preference for the North conformation were more refractory to excision by both WT-RT and AZTr-RT. We found that DNA primers that contain North puckered-nucleotides at the 3′-terminus can also affect the translocation status of the RT/template/primer complex, which provides an underlying mechanism to avoid being excised. Together, these results point to a correlation between the sugar conformation of the 3′-terminal nucleotide, the precise position of HIV-1 RT on its nucleic acid substrate, and, in turn, its catalytic function. Nucleotide sugar conformation is therefore an important parameter in defining the susceptibility to RT-catalyzed phosphorolytic excision.
K. Yamada, A.S. Wahba, J. Bernatchez, T. Ilina, S. Martinez-Montero, M. Habibian, G.F. Deleavey, M. Gotte, M.A. Parniak, and M.J. Damha† (2015), ACS Chemical Biology
You can view the full article here.
Locked 2'-diF-rU Modified Nucleic Acids
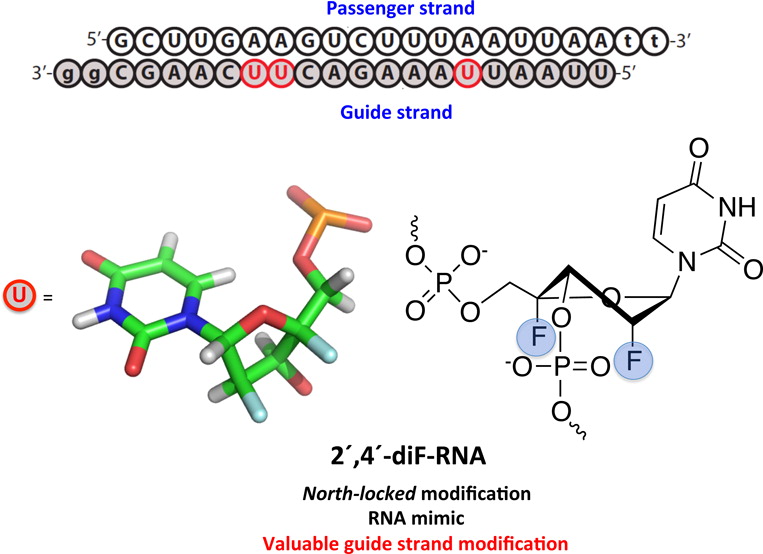
2′-Deoxy-2′,4′-difluorouridine (2′,4′-diF-rU) was readily incorporated into DNA and RNA oligonucleotides via standard solid phase synthesis protocols. NMR and thermal denaturation (Tm) data of duplexes was consistent with the 2′,4′-diF-rU nucleotides adopting a rigid North (RNA-like) sugar conformation, as previously observed for the nucleoside monomer. The impact of this modification on Tm is neutral when incorporated within RNA:RNA duplexes, mildly destabilizing when located in the RNA strand of a DNA:RNA duplex, and highly destabilizing when inserted in the DNA strand of DNA:RNA and DNA:DNA duplexes. Molecular dynamics calculations suggest that the destabilization effect in DNA:DNA and DNA:RNA duplexes is the result of structural distortions created by A/B junctions within the helical structures. Quantum mechanics calculations suggest that the “neutral” effect imparted to A-form duplexes is caused by alterations in charge distribution that compensate the stabilizing effect expected for a pure North-puckered furanose sugar. 2′,4′-diF-RNA modified siRNAs were able to trigger RNA interference with excellent efficiency. Of note, incorporation of a few 2′,4′-diF-rU residues in the middle of the guide (antisense) strand afforded siRNAs that were more potent than the corresponding siRNAs containing LNA and 2′-F-ANA modifications, and as active as the 2′-F-RNA modified siRNAs.
S. Martinez-Montero, G.F. Deleavey, N. Martin-Pintado, J. Fakhoury, C. Gonzalez, and Masad J. Damha† (2015) ACS Chemical Biology
You can view the full article here.
Synthesis and Properties of 2',4'-diFANA Modified Nucleic Acids
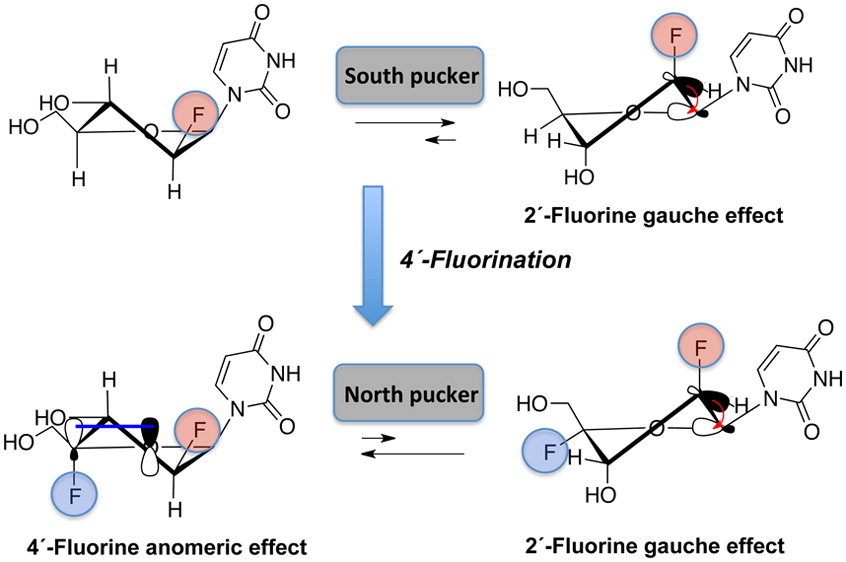
We report the synthesis, thermal stability, and RNase H substrate activity of 2′-deoxy-2′,4′-difluoroarabino-modified nucleic acids. 2′-Deoxy-2′,4′-difluoroarabinouridine (2,′4′-diF-araU) was prepared in a stereoselective way in six steps from 2′-deoxy-2′-fluoroarabinouridine (2′-F-araU). NMR analysis and quantum mechanical calculations at the nucleoside level reveal that introduction of 4′-fluorine introduces a strong bias toward the North conformation, despite the presence of the 2′-βF, which generally steers the sugar pucker toward the South/East conformation. Incorporation of the novel monomer into DNA results on a neutral to slightly stabilizing thermal effect on DNA–RNA hybrids. Insertion of 2′,4′-diF-araU nucleotides in the DNA strand of a DNA–RNA hybrid decreases the rate of both human and HIV reverse transcriptase-associated RNase H-mediated cleavage of the complement RNA strand compared to that for an all-DNA strand or a DNA strand containing the corresponding 2′-F-araU nucleotide units, consistent with the notion that a 4′-fluorine in 2′-F-araU switches the preferred sugar conformation from DNA-like (South/East) to RNA-like (North).
S. Martinez-Montero, G. Deleavey, A. Dierker-Viik, P. Lindovska, T. Ilina, G. Portella, M. Orozco, M.A. Parniak, C. Gonzalez, and M.J. Damha† (2015), Journal of Organic Chemistry, 80 (6): 3083-3091.
You can view the full article here.
Synthesis, Structure, and Conformational Analysis of Six-Membered 1,3-Oxathiane Nucleoside Analogues
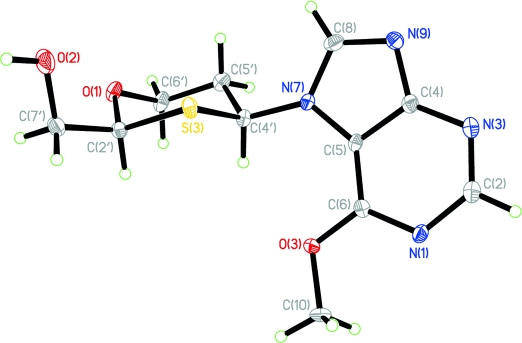
We report on the synthesis, characterization, and conformational analysis of 1,3-oxathiane nucleosides of both pyrimidine and purine nucleobases. The novel nucleoside analogues were prepared from readily available starting materials as α/β anomeric mixtures. The purine nucleosides were obtained as separable N7/N9 regioisomers. The structures were identified by extensive NMR analysis. The X-ray structure of an N7-β-OMe-purine nucleoside analogue shows that the 1,3-oxathiane sugar ring adopts a perfect chair conformation with both the hydroxymethyl and the nucleobase placed in equatorial positions.
Abou Assi, H.; Martinez-Montero, S.; Dixit, D.; Chua, Z.; Bohle, D.S.; Damha, M.J. (2015). Eur. J. Org. Chem. 1945-1953.
View the full article here.
NAR Breakthrough! Structural Basis of Lariat RNA Recognition by the Intron Debranching Enzyme Dbr1
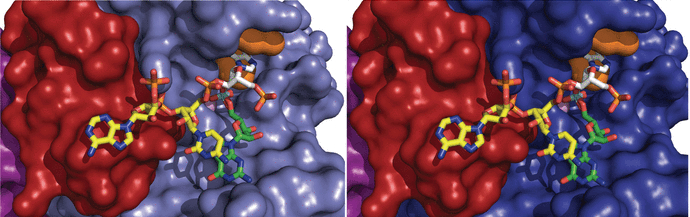
The enzymatic processing of cellular RNA molecules requires selective recognition of unique chemical and topological features. The unusual 2',5'-phosphodiester linkages in RNA lariats produced by the spliceosome must be hydrolyzed by the intron debranching enzyme (Dbr1) before they can be metabolized or processed into essential cellular factors, such as snoRNA and miRNA. Dbr1 is also involved in the propagation of retrotransposons and retroviruses, although the precise role played by the enzyme in these processes is poorly understood. Here, we report the first structures of Dbr1 alone and in complex with several synthetic RNA compounds that mimic the branchpoint in lariat RNA. The structures, together with functional data on Dbr1 variants, reveal the molecular basis for 2',5'-phosphodiester recognition and explain why the enzyme lacks activity toward 3',5'-phosphodiester linkages. The findings illuminate structure/function relationships in a unique enzyme that is central to eukaryotic RNA metabolism and set the stage for the rational design of inhibitors that may represent novel therapeutic agents to treat retroviral infections and neurodegenerative disease.
E.J. Montemayor, A. Katolik, A.B. Taylor, J. Scheurmann, D.J. Combs, R. Johnsson, S.P. Holloway, S.W. Stevens†, M.J. Damha†, and P.J. Hart† (2014) Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1, Nucleic Acids Research ("Breakthrough Article")
View the full article here.
